Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis
Figures
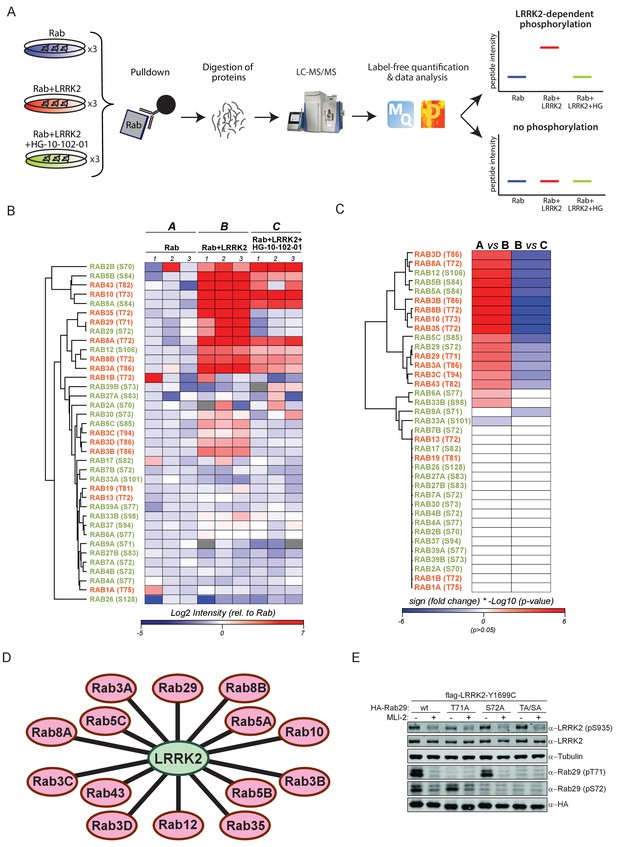
14 Rab GTPases are phosphorylated by LRRK2 when overexpressed HEK293s.
(A) Workflow for identifying Rabs that are phosphorylated by LRRK2 in an overexpression system. The individual Rab proteins were overexpressed alone or expressed in combination with active (G2019S)- or chemically inhibited LRRK2 (HG-10-102-01, 3 µM, 3 hr, n = 3). The Rabs were then affinity-enriched, digested and analyzed by label-free LC/MS-MS. Mass spectra database search and data analysis were performed using MaxQuant and Perseus, respectively. Substrates were scored when the predicted Rab phosphopeptide was detected, upregulated upon LRRK2 expression and reduced after inhibitor treatment. (B) Heat map of phosphopeptide intensities (Log2) and (C) Tukey adjusted p-values (-Log10, multiplied by the sign of the fold change of the respective group comparison [A = Rab, B = Rab + LRRK2, C = Rab + LRRK2 +HG-10-102-10]). Values missing in all replicates of one group were imputed and non-imputed, missing values are in grey. (D) Scheme of the 14 Rab proteins that are phosphorylated by LRRK2. (E) Western blot confirming that both Rab29-pT71 and Rab29-pS72 are phosphorylated by LRRK2-Y1699C when overexpressed in HEK293 cells.

Sequence alignment of 50 Rabs in which the predicted LRRK2 phosphorylation site is conserved.
https://doi.org/10.7554/eLife.31012.003
Sequence alignment of Rab10, Rab38 and Rab32 and western blot analysis of 52 overexpressed Rab GTPases.
(A) Sequence alignment showing that the equivalent site to Rab10-T73 is not conserved in Rab32 and Rab38. (B) Western blot analysis of 52 epitope-tagged Rab proteins expressed alone or in combination with LRRK2-G2019S (n = 3). HG-10-10-102 (3 µM, 3 hr) was used to inhibit LRRK2.

Both Rab29-T71 and Rab29-S72 are phosphorylated by overexpressed LRRK2 in HEK293 cells.
(A) Western blot of HA-Rab29-S72A and (B) HA-Rab29-T71A expression in HEK293 cells.(C) MS-quantified Rab29-pT71 and (D) Rab29-pS72 phosphopeptide intensities of overexpressed proteins in HEK293 cells.
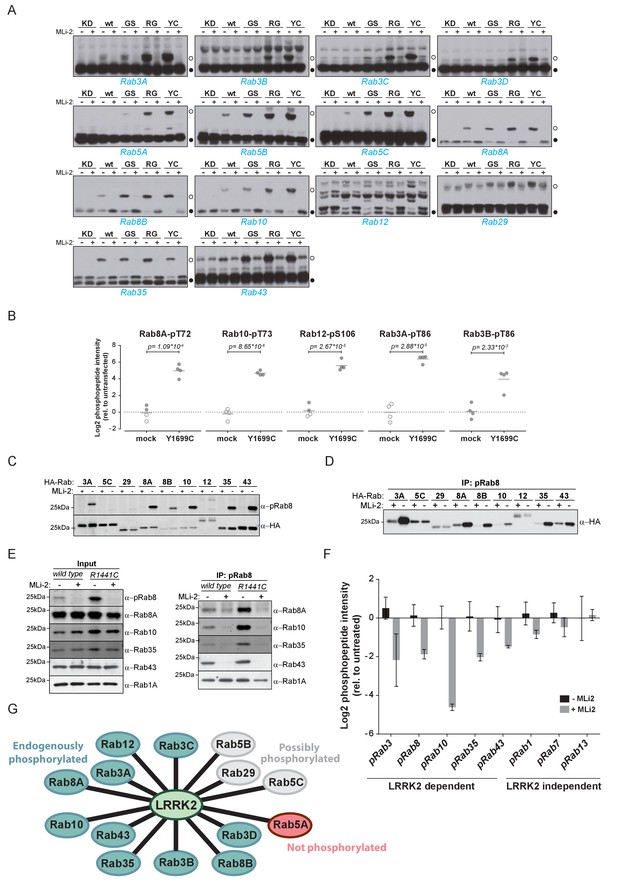
Ten endogenous Rabs are phosphorylated by LRRK2 in cells.
(A) Phos-tag SDS-PAGE of 14 Rabs co-expressed with different LRRK2 variants (KD = kinase dead, wt = wild type, GS= G2019S, RG = R1441G, YC= Y1699C) in HEK293 cells. Filled circles indicate unmodified Rabs and open circles, phosphorylated forms (MLi-2 = 100 nM, 1 hr). (B) MS-quantified Rab3A-pT86, Rab3B-pT86, Rab8A-pT72, Rab10-pT73 and Rab12-pS106 peptide intensities from mock and LRRK2-Y1699C transfected cells. Open circles indicate imputed values. (C) Western blot analysis of HA-tagged Rabs co-expressed with LRRK2-Y1699C in HEK293 cells (-/+100 nM MLi-2, 2 hr) using polyclonal anti-phospho-Rab8 and HA antibodies. (D) Same as (C) but Rabs were immunoprecipitated using the anti-pRab8 antibody and HA antibody was used for detection. (E) Western blot of endogenous Rabs in wild type and LRRK2-R1441C MEFs (-/+100 nM MLi-2, 1 hr) using the indicated antibodies (left) and immunoprecipitation of Rabs using anti-pRab8 antibody (right). (F) MS-quantified Rab phosphosite intensities of pRab8 immunoprecipitation in R1441C MEFs (-/+100 nM MLi-2, 2 hr). Error bars are mean ±SEM (n = 3). (G) 10 Rab proteins at endogenous expression levels are phosphorylated by LRRK2 in cells (green). Rab5A is not (red) and Rab5B, Rab5C and Rab29 are possibly phosphorylated (grey).
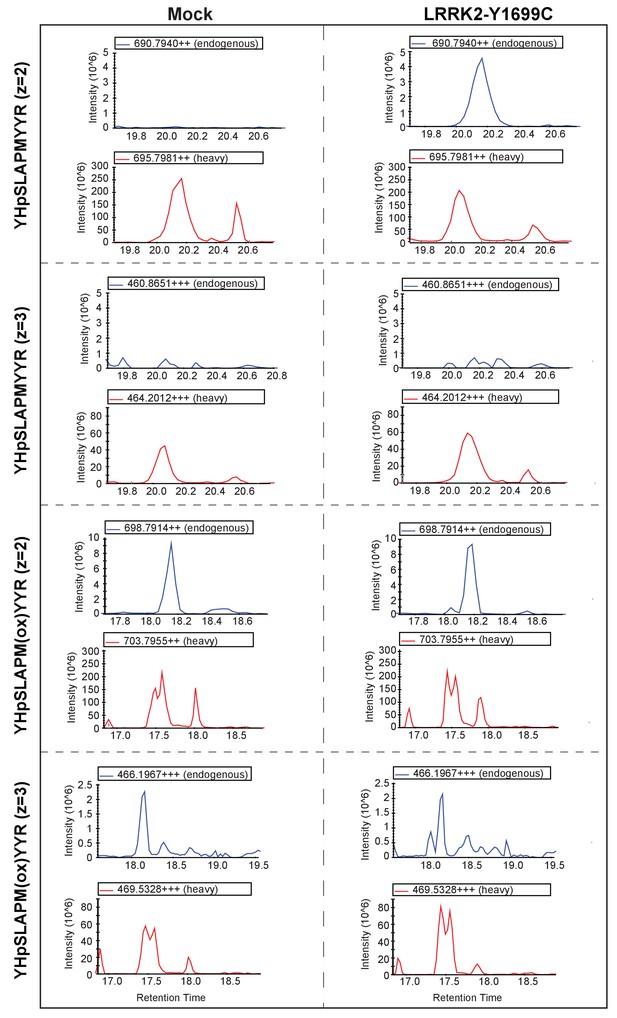
Rab5A was immunoprecipitated from mock or LRRK2-Y1699C-transfected HEK293 cells.
Before LC-MS/MS analysis, 300 fmol of a stable isotope-labeled peptide corresponding to the endogenous, predicted LRRK2 target peptide was spiked in the sample. Both light and heavy counterpart peptides were recorded simultaneously using multiplexed SIM scans (see Materials and methods). Extracted ion chromatograms (XICs) of doubly (z = 2)- and triply (z = 3)-charged oxidized and non-oxidized YHpSLAPMYYR ions are shown.
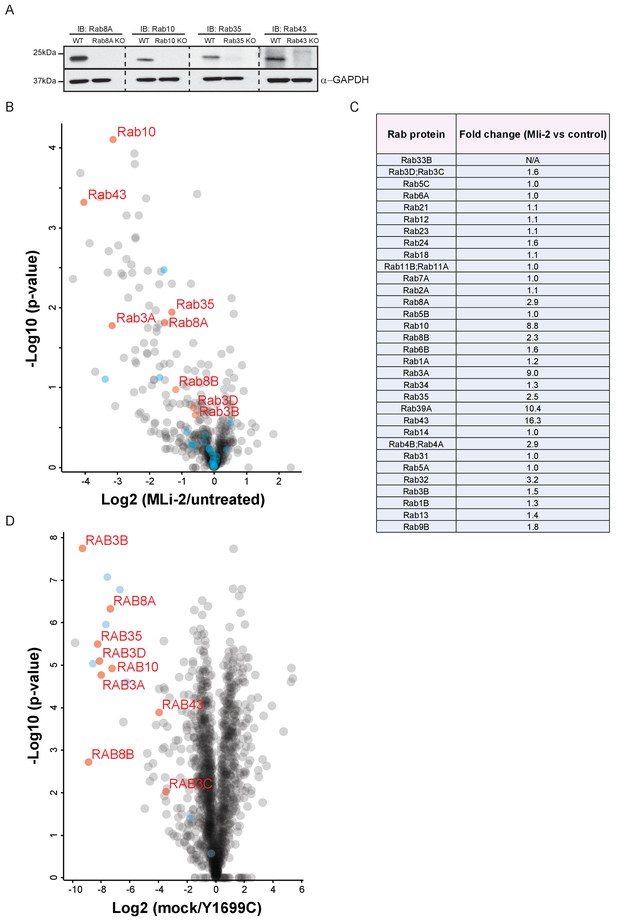
Nine Rab GTPases containing a Thr site in their switch-II domain are phosphorylated by LRRK2.
(A) Western blot analysis of A549 CRISPR-CAS9 knockout cells demonstrating specificity of anti-Rab8A, Rab10, Rab35 and Rab43 antibodies. (B) Volcano plot summarizing the AE-MS analysis of LRRK2-R1441C MEFs, left untreated or treated with MLi-2 (100 nM) for 2 hr. (C) Table of 32 quantified Rabs with fold-changes (MLi-2 vs untreated) identified in (B). (D) Volcano plot of pRab8 immunoprecipitation in mock or LRRK2-Y1699C transfected HEK293 cells. Rab proteins that were identified in our initial, overexpression screen are in red while other Rab family members are in light blue.
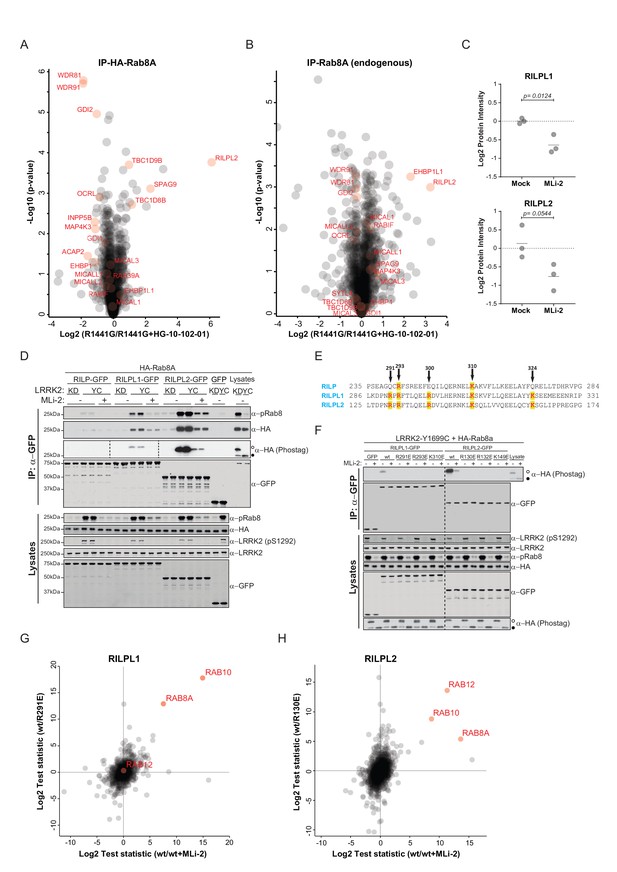
Phosphorylation-specific protein binding to Rabs.
(A) AE-MS of HA-Rab8A and (B) endogenous Rab8A using extracts of GFP-LRRK2-R1441G expressing Flp-In T-Rex HEK293 cells. Expression of the kinase was induced for 48 hr by doxycycline (1 µg/ml) and LRRK2 inhibited using HG-10-102-01 (3 µM, 3 hr). LRRK2-regulated, known and unknown Rab8A-binding proteins in both AE-MS experiments are highlighted in red. (C) Label-free (LFQ [Cox et al., 2014]), MS-quantified RILPL1 and RILPL2 levels after immunoprecipitation of Rab8A from mock- or MLi-2 (200 nM, 2 hr) treated LRRK2-R1441C MEFs. (D) Pulldown of GFP-tagged RILP, RILPL1 or RILPL2, transiently expressed with HA-Rab8A and the indicated LRRK2 variants (KD= Y1699C/D2017A) in HEK293 cells. Western blot after Phos-tag SDS-PAGE was used to detect interacting proteins using the indicated antibodies. (MLi-2 = 150 nM, 2 hr). (E) Sequence alignment of the RILP homology (RH) domains of RILP, RILPL1 and RILPL2 showing five conserved basic residues, which are highlighted. (F) Same as (D) but using different RILPL1 and RILPL2 mutants. For phos-tag blots, filled circles indicate non phosphorylated proteins and open circles phosphorylated proteins. (G) AE-MS of GFP-RILPL1 (wt or R291E) and (H) GFP-RILPL2 (wt or R130E), expressed with LRRK2-Y1699C in HEK293 cells, and treated or not with MLi-2 (for wt, 200 nM, 2 hr). The student’s two sample test statistic (Log2) of the indicated comparisons was used for plotting.
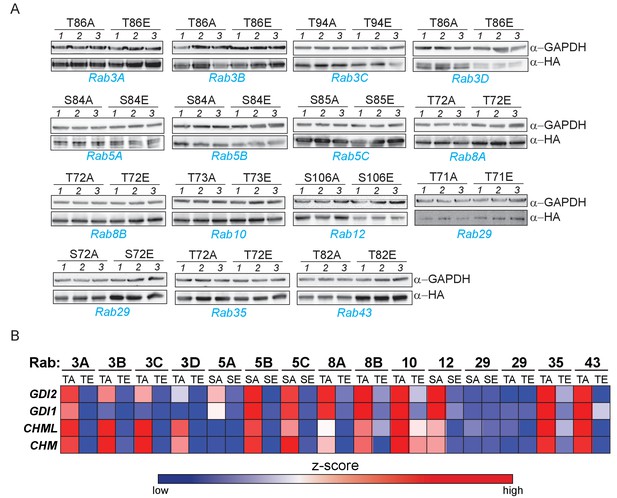
Phosphomimetic S/T->E mutation of the LRRK2 phosphorylation site abrogates GDI1/2 and CHM/CHML binding in 13 Rab GTPases.
(A) Western blot analysis of 15 S/T→A or S/T→E mutants expressed in triplicate in HEK293 cells. (B) Heat map illustrating binding of GDI1/2 and CHM/CHML to 30 different Rab S/T→A or S/T→E mutants, which were determined by AE-MS. Z-scored, median LFQ intensities are plotted (Cox et al., 2014).
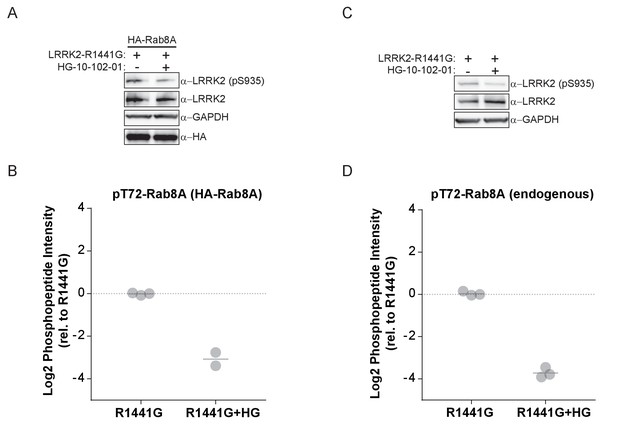
MS-quantified pT72-Rab8A peptide levels in HEK293 cells expressing LRRK2-R1441G.
(A) Western blot analysis of HA-Rab8A and GFP-LRRK2-R1441G expression and (B) MS-quantified pT72-Rab8A levels. (C, D) Same as (A, B) but with endogenous Rab8A expression levels. HG-10-102-01 treatment was for 3 hr (3 µM) and kinase expression induced by doxycycline for 48 hr (1 µg/ml).

Phospho-dependent interaction of Rab8A, Rab10 and Rab12 with RILPL1/2.
(A) GFP pulldown of different mutants of the C-terminal domain of RILPL1 (280-END) transiently expressed with HA-Rab8A and LRRK2-Y1699C in HEK293 cells (wt = wild type). Western blot and Phos-tag were used to detect interacting proteins using the indicated antibodies. (MLi-2 = 150 nM, 2 hr). (B), (C) and (D) Same as (A) but using full-length GFP-tagged RILP, RILPL1 or RILP2, co-expressed with either Rab8A, Rab10 or Rab12 and LRRK2-Y199C. For phos-tag blots filled circles indicate non-phosphorylated proteins and open circles phosphorylated proteins.
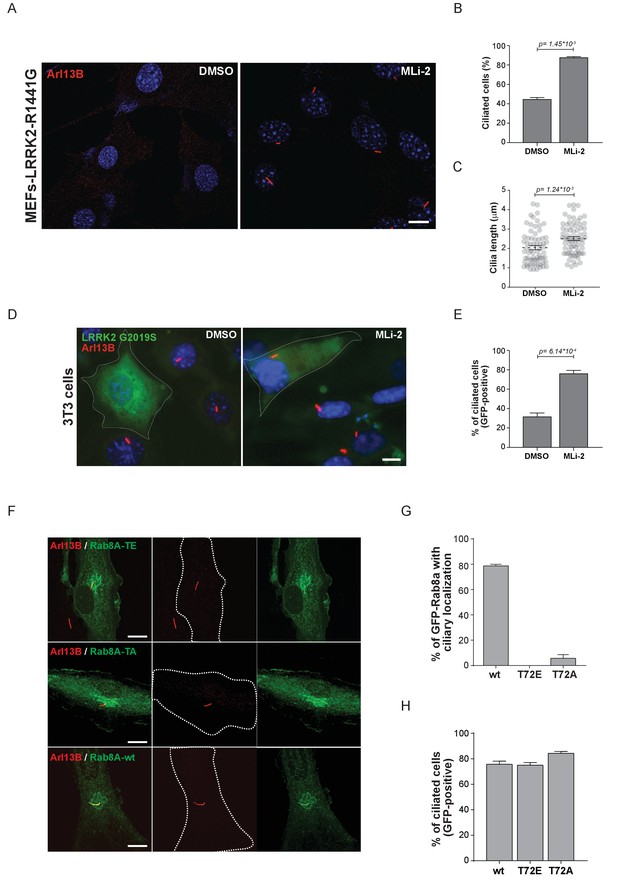
Pathogenic LRRK2 mutations inhibit primary cilia formation.
(A) LRRK2-R1441G knock-in MEFs were serum starved overnight and treated with 200 nM MLi-2 (right) or DMSO (left). Primary cilia were stained using mouse anti-Arl13B (red) and nuclei using DAPI. (B) Quantification of primary cilia (Arl13B staining, n = 2,>100 cells per condition) and (C) cilia length (70 per condition). Scale bar = 10 µm. (D) NIH3T3 cells transfected with eGFP-LRRK2-G2019S were serum starved for 24 hr in the presence or absence of MLi-2 (200 nM). Scale bar = 10 µm. (E) Quantification of primary cilia (Arl13B positive) in GFP-positive cells;>50 cells per replicate were counted (n = 3). Error bars represent SEM and p-values were determined using unpaired, two-tailed Student’s t-tests. (F) RPE-1 cells were infected with a lentivirus encoding GFP-Rab8A (T72E, T72A or wt) and 48 hr later, serum starved overnight to induce ciliation; primary cilia were detected with mouse anti-Arl13B (red). Dotted lines indicate cell outlines. Scale bars = 10 µm. (G) Quantification of cells with ciliary (Arl13B positive) GFP-Rab8A localization; (H) Quantitation of primary cilia (Arl13B staining) in GFP-positive cells. Error bars represent SEM from three experiments with >75 cells counted per condition.
Additional files
-
Supplementary file 1
Summary of used proteases and identified peptides for the proteomic analysis of 52 Rab GTPases.
Proteomics raw data have been deposited to the ProteomeXchange Consortium (Vizcaíno et al., 2014) via the PRIDE partner repository with the data set identifier PXD007214.
- https://doi.org/10.7554/eLife.31012.014
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31012.015





